Frontiers in Technology
Poster Session B
(1131-B) Harnessing and Validating Deep Learning-based Image Analysis for Cell Painting as an Unbiased Approach to Phenotypic Drug Discovery
Wednesday, May 29, 2024
10:30 - 11:15 CEST
Location: Exhibit Hall

Victor Wong, PhD
Chief Scientific Officer
Core Life Analytics
`S-Hertogenbosch, Noord-Brabant, Netherlands
Poster Presenter(s)
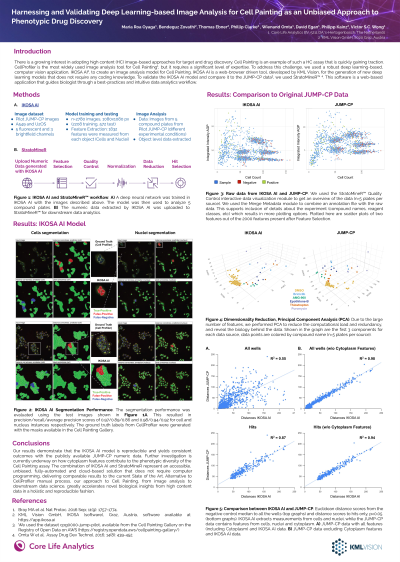
Abstract: There is a growing interest in adopting high content (HC) image-based approaches for target and drug discovery. Cell Painting (CP) is an example of such a HC assay that is quickly gaining traction. CellProfiler is the most widely used image analysis tool for CP, but it requires a significant level of expertise. To address this challenge, we used a robust deep learning-based, computer vision application, IKOSA AI, to create an image analysis model for CP. IKOSA AI is a web-browser driven tool, developed by KML Vision, for the generation of new deep learning models that does not require any coding knowledge.
Our model was developed through the following steps: we generated ground truth labels from the CP Gallery JUMP-CP pilot dataset. In order to maximize the diversity of the phenotypes, the training database contains A549 and U2OS cell lines, treated with various chemical and genetic perturbations. The database was split into training (n=2,208 images) and validation (n=572 images), each image has 5 fluorescent and 3 brightfield channels. A deep neural network was subsequently trained using IKOSA AI and the segmentation performance was evaluated on the validation images. This resulted in precision/recall/average precision scores of 0.97/0.89/0.86 and 0.98/0.94/0.92 for cell and nucleus instances respectively. Based on the instance segmentation outputs, 1,832 features were extracted from each object. We further applied StratoMineR to compare IKOSA AI cell level data with the JUMP-CP dataset, using 5 compound plates with various experimental conditions. One of the barriers to analyzing this data is the vast amount of features, therefore, we used Spearman's correlation to exclude redundant features. We used PCA to generate 8 components and performed hit selection based on the Euclidean distance.
The IKOSA AI platform extracts measurements from cells and nuclei, while the JUMP-CP data contains features from cells, nuclei and cytoplasm. We compared the Euclidean distances of all compounds from both datasets and found a correlation coefficient of 0.55. We subsequently found 315 and 304 compounds that were significantly different from DMSO (p-value < 0.05) from the JUMP-CP and IKOSA datasets, respectively. Within those hits, 294 were common hits. Interestingly, when cytoplasm features were excluded from the JUMP-CP data alone, the correlation between IKOSA and JUMP-CP was 0.98. We also found 295 common hits between JUMP-CP and IKOSA datasets.
Our results demonstrate that the IKOSA AI model is reproducible and yields consistent outcomes from the original JUMP-CP data. Further investigation is currently underway on how cytoplasm features contribute to the phenotypic diversity of the CP assay. The combination of IKOSA AI and StratoMineR represent an accessible, unbiased, fully-automated solution that does not require computer programming, delivering comparable results to the current State of the Art. Alternative to the manual process, our approach to CP from image analysis to downstream data science greatly accelerates novel biological insights from high content data in a holistic and reproducible fashion.
Our model was developed through the following steps: we generated ground truth labels from the CP Gallery JUMP-CP pilot dataset. In order to maximize the diversity of the phenotypes, the training database contains A549 and U2OS cell lines, treated with various chemical and genetic perturbations. The database was split into training (n=2,208 images) and validation (n=572 images), each image has 5 fluorescent and 3 brightfield channels. A deep neural network was subsequently trained using IKOSA AI and the segmentation performance was evaluated on the validation images. This resulted in precision/recall/average precision scores of 0.97/0.89/0.86 and 0.98/0.94/0.92 for cell and nucleus instances respectively. Based on the instance segmentation outputs, 1,832 features were extracted from each object. We further applied StratoMineR to compare IKOSA AI cell level data with the JUMP-CP dataset, using 5 compound plates with various experimental conditions. One of the barriers to analyzing this data is the vast amount of features, therefore, we used Spearman's correlation to exclude redundant features. We used PCA to generate 8 components and performed hit selection based on the Euclidean distance.
The IKOSA AI platform extracts measurements from cells and nuclei, while the JUMP-CP data contains features from cells, nuclei and cytoplasm. We compared the Euclidean distances of all compounds from both datasets and found a correlation coefficient of 0.55. We subsequently found 315 and 304 compounds that were significantly different from DMSO (p-value < 0.05) from the JUMP-CP and IKOSA datasets, respectively. Within those hits, 294 were common hits. Interestingly, when cytoplasm features were excluded from the JUMP-CP data alone, the correlation between IKOSA and JUMP-CP was 0.98. We also found 295 common hits between JUMP-CP and IKOSA datasets.
Our results demonstrate that the IKOSA AI model is reproducible and yields consistent outcomes from the original JUMP-CP data. Further investigation is currently underway on how cytoplasm features contribute to the phenotypic diversity of the CP assay. The combination of IKOSA AI and StratoMineR represent an accessible, unbiased, fully-automated solution that does not require computer programming, delivering comparable results to the current State of the Art. Alternative to the manual process, our approach to CP from image analysis to downstream data science greatly accelerates novel biological insights from high content data in a holistic and reproducible fashion.
