Frontiers in Technology
Poster Session B
(1141-B) Multiplex Screening of DNA-Encoded Small Molecule Libraries inside a Living Cell
Wednesday, May 29, 2024
10:30 - 11:15 CEST
Location: Exhibit Hall

Lars Kolster Petersen, PhD (he/him/his)
Associate Director Chemistry
Vipergen ApS
Copenhagen V, Hovedstaden, Denmark
Poster Presenter(s)
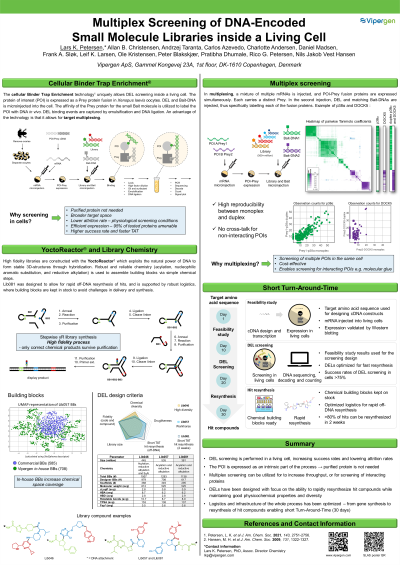
Abstract: Screening of DNA-encoded libraries (DELs) has gained momentum as an important hit identification technique over the last two decades. In contrast to high throughput-screening, DNA barcoding of molecules in conjunction with next-generation sequencing allows for a deeper and more cost-effective probing of chemical space. Recently, we published cellular binder trap enrichment (cBTE) technology for DEL screening.(1) Target proteins are expressed and screened inside oocytes from the South African clawed frog Xenopus laevis, providing screening conditions of high physiological relevance and simultaneously circumventing any demand for highly purified target protein. The oocyte cell system was chosen as it is a readily available cellular system which is well-characterized in terms of handling and expression of non-endogenous proteins. Introduction of both mRNA and DEL into the cytosol was done by microinjection, thereby resolving the fundamental problem of delivering DELs across cell membranes for in vivo screening. The technology offers a high level of convenience as only the amino acid sequence of the target protein is required for mRNA construction. Also, it allows for screening of high numbers of small molecules offered by DELs (~500 million library members), thus providing easy access to novel chemical matter for target proteins which are not amenable to conventional protein expression. In addition, simultaneous screening of more than one target (multiplexing) in the same cell has been demonstrated. Multiplexing provides an important tool for identification of hits against larger protein complexes and biological systems with multiple interaction partners. Design criteria and construction of DELs will be presented and examples of results from screening in multiplex will be discussed.
(1) Petersen, L. K et al J. Am. Chem. Soc. 2021, 143, 2751-2756
(1) Petersen, L. K et al J. Am. Chem. Soc. 2021, 143, 2751-2756
