Frontiers in Technology
Poster Session A
(1150-A) Screening for RAS inhibitor combination targets using a multiplexed CRISPR platform that incorporates drug sensitivity profiling, mode of action assays and cell line generation
Tuesday, May 28, 2024
16:30 - 17:15 CEST
Location: Exhibit Hall

Charlie Dunlop, Senior Scientist
Dr
AstraZeneca
CAMBRIDGE, England, United Kingdom
Poster Presenter(s)
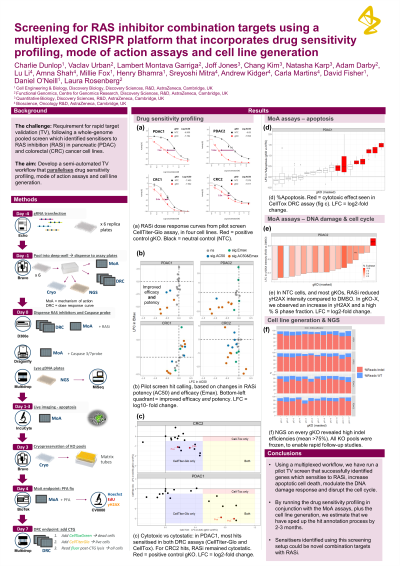
Abstract: The AstraZeneca CRISPR pipeline for target discovery uses arrayed CRISPR screening with synthetic guide RNAs to validate hits from lentiviral whole-genome pooled CRISPR screens. Since arrayed workflows can be time-consuming and laborious, it is favourable to maximise the information obtained from one screen, as this enables detailed annotation and faster triaging of hits. For drug-target interaction studies, it is common to run cell proliferation and mode of action (MoA) assays sequentially. Parallel execution could significantly reduce target validation timelines.
Following a whole-genome pooled screen which identified sensitisers to RAS inhibition (RASi) in pancreatic and colorectal cancer cell lines, we established a semi-automated, arrayed KO workflow that features multiple readouts and thus extensively annotates hits for target validation. Each gene knockout (gKO) was profiled in a 7-point dose response assay quantifying changes in RASi potency and efficacy, plus assessment of cytostatic and cytotoxic effects. In parallel, we used live cell imaging to capture apoptosis as a mode of cell death and an immunofluorescence assay to monitor both cell cycle changes and induction of DNA damage. Additionally, next-generation amplicon sequencing of every gKO was applied to validate editing efficiency and to link the observed phenotypes to genotype. Furthermore, we streamlined follow-up studies of top hits by cryopreservation of every KO pool.
We developed this multiplexed workflow by employing several automated liquid handlers, and confirmed the full protocol’s handling and statistical robustness in a pilot screen. Data from the primary hit-ranking assay, CellTiter-Glo, showed that automation produced low plate-to-plate variability (neutral control average CV = 6.7%) and a wide assay window (up to 13.4-fold shift in IC50 using a positive control gKO). The mechanistic imaging readouts also demonstrated good assay windows, and our NGS assessments revealed an average CRISPR editing efficiency of over 75% in the cryopreserved KO pools.
By running the drug sensitivity profiling in conjunction with the MoA assays, plus the cell line generation, we estimate that we have sped up the hit annotation process by 2-3 months. We have used the workflow to validate top hits from a RASi pooled screen, successfully identifying genes which sensitise to RASi across cell lines, increase apoptotic cell death, modulate the DNA damage response and disrupt the cell cycle. Sensitisers identified using this screening setup could be novel combination targets with RASi. In summary, by utilising automation and running numerous assays in parallel, our multiplexed arrayed CRISPR KO platform can significantly expedite hit-triaging and will bolster the AstraZeneca CRISPR pipeline for target discovery.
Following a whole-genome pooled screen which identified sensitisers to RAS inhibition (RASi) in pancreatic and colorectal cancer cell lines, we established a semi-automated, arrayed KO workflow that features multiple readouts and thus extensively annotates hits for target validation. Each gene knockout (gKO) was profiled in a 7-point dose response assay quantifying changes in RASi potency and efficacy, plus assessment of cytostatic and cytotoxic effects. In parallel, we used live cell imaging to capture apoptosis as a mode of cell death and an immunofluorescence assay to monitor both cell cycle changes and induction of DNA damage. Additionally, next-generation amplicon sequencing of every gKO was applied to validate editing efficiency and to link the observed phenotypes to genotype. Furthermore, we streamlined follow-up studies of top hits by cryopreservation of every KO pool.
We developed this multiplexed workflow by employing several automated liquid handlers, and confirmed the full protocol’s handling and statistical robustness in a pilot screen. Data from the primary hit-ranking assay, CellTiter-Glo, showed that automation produced low plate-to-plate variability (neutral control average CV = 6.7%) and a wide assay window (up to 13.4-fold shift in IC50 using a positive control gKO). The mechanistic imaging readouts also demonstrated good assay windows, and our NGS assessments revealed an average CRISPR editing efficiency of over 75% in the cryopreserved KO pools.
By running the drug sensitivity profiling in conjunction with the MoA assays, plus the cell line generation, we estimate that we have sped up the hit annotation process by 2-3 months. We have used the workflow to validate top hits from a RASi pooled screen, successfully identifying genes which sensitise to RASi across cell lines, increase apoptotic cell death, modulate the DNA damage response and disrupt the cell cycle. Sensitisers identified using this screening setup could be novel combination targets with RASi. In summary, by utilising automation and running numerous assays in parallel, our multiplexed arrayed CRISPR KO platform can significantly expedite hit-triaging and will bolster the AstraZeneca CRISPR pipeline for target discovery.
